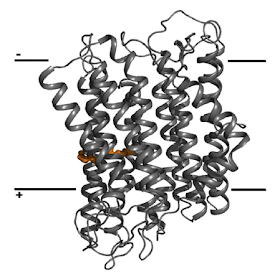
So this is the predicted structure of the modern MrpA-like functional antiporter from N thermophilus, nt-Nha:
courtesy of
A missing link between complex I and group 4 membrane-bound [NiFe] hydrogenases
As you can see it is a double channel and it is homologous to NouL from mammalian (and others) complex I. The two marked amino acids are a glutamic acid and a lysine which are conserved in NuoL. I would guess that the left hand channel is the proton channel and the right hand one is for the antiported Na+ ion. In the modern bacterium N thermophilus this antiporter is electrogenic, ie it moves more than one H+ inwards for each Na+ outwards. This may or may not have been the case in LUCA.
So nt-Nha looks a lot like NuoL and so quite like NuoM and NuoN (and MrpA, MrpD and MbhH) but it is the only one to antiport. The antiport mechanism really hasn't been worked out yet.
I guess the next thing to look at is from the same paper. This time we're looking at NuoH, the anchor point at the base of all complex I family of proton pumps. Here it is as a model with NuoH in gold superimposed over the "left hand" channel of NuoL in grey:
Half of NuoL is structurally pretty well identical to NuoH.
From the proto-Ech posts I consider NuoH to be derived from the primordial channel designed to allow oceanic pH penetration in towards the NiFe catalytic site (at alkaline hydrothermal vent pH) to generate reduced ferredoxin as the core power molecule of early LUCA, able to reduce CO2 to CO at the CODH/ACS complex.
So a derivative of the ancestral proton channel formed half of NuoL. I think it is very likely that this is the case for all of the "antiporter-like" subcomplexes, though clearly some changes in function have occurred. What made up the other channel of the ancestral antiporter? For a clue as to this one we have to change subunit and change nomenclature. The work has not been done for NuoL but it has been done for the membrane bound hydrogenase of P furiosus (subunit MbhH) which is structurally more closely related to NuoN rather than NuoL.
Let's switch papers and go back to
Structure of an Ancient Respiratory System
Here we are viewing subunit MbhH, remember that we think that the left hand channel is probably derived from the ancestor of NuoH:
Now, here's the clever bit. If we draw a horizontal line across the left hand channel, say just below the "9", we can rotate the channel around this line so all of the labels 9, 10, 11, 12 and 13 end up on the bottom far side of the model. With this done the two channels are superimposable, rather neatly:
So the right hand side of MbhH is simply the inverse of the left hand side.
We only need one protein, the original ancestral proton channel, to make both sides of the original ancestral antiporter. Minor modification would allow one side for H+ and the other for Na+. Then some sort of coupling to convert H+ in to drive Na+ out. But basically we can make all of the modern "antiporter-like" subunits from an ancestral derivative of two conjoined NuoH-like channels.
That is so neat it has to be correct. So it's probably wrong!
That coupling process might be extractable from work being done with modern complex I but I have only skim read the paper so far, so I'm not sure how much further I can progress the current fairy tale.
Peter
Wednesday, March 27, 2019
Tuesday, March 26, 2019
Life (26) MrpA MrpD NuoL NuoM and NuoN. Plus nt-Nha.
A few years ago I mentioned this paper
Homologous protein subunits from Escherichia coli NADH:quinone oxidoreductase can functionally replace MrpA and MrpD in Bacillus subtilis
In brief they had Bacillus subtilis strains with either an MrpA knockout or an MrpD knockout. The E coli complex I equivalent of NuoL can replace MrpA and the NuoN equivalent can replace MrpD in B subtilis. NouM doesn't seem do either. But all five subunits look very similar to each other and all are clearly related. NuoL, NuoM and NuoN are always described as "antiporter-like". MrpA and MrpD are thought to be antiporters but none ever work alone, the whole complex is needed, so they are probably "antiporter-like" too. They all appear to have been derived from an antiporter but any intrinsic antiporting has been lost.
Which makes me sad because it seems very probable that all of the above subunits are derived from the primordial antiporter at the origin of life which initiated Na+ bioenergetics and all that followed on from that.
Then came Natranaerobius thermophilus. N thermophilus is not really in the league of P furiosus, it's okay growing at up to 57 degC (which will still scald you) but no higher and it has adapted its membrane to remain proton tight at this temperature. BTW it's strictly anaerobic, is an halophile and an alkaliphile. It has (among several) one family of modern antiporters which are clearly genetically related to the MrpA of modern Clostridium tetani (and probably all other MrpAs). Modern nt-Nha is a fully functional antiporter as a stand-alone single gene protein. As these folks say:
The halophilic alkalithermophile Natranaerobius thermophilus adapts to multiple environmental extremes using a large repertoire of Na+(K+)/H+ antiporters
"Gene nt-Nha had 35% identity to the shaA (mrPA) gene of Clostridium tetani. The Mrp proteins belong to the monovalent cation/proton antiporter-3 protein family. This family is composed of multi-component Na+/H+ and K+/H+ antiporters encoded by operons of six or seven genes, and all genes are required for full function in Na+ and alkali resistance (Ito et al., 2000). Sequence analysis of the
regions surrounding gene nt-Nha, however, did not show that it was part of an operon. This indicates that gene nt-Nha does not encode a subunit of an Mrp system, but rather a mono-subunit antiporter".
EDIT number 2: These people have isolated an MrpA from an archaeal species which will antiport on its own, which makes it very similar to nt-Nha. There is also some evidence that complex I can function as a partial Na+/H+ antiporter as in this paper. NuoL is the main suspect. END EDIT.
Neither the MrpA-like nt-Nha nor the modern MrpA subunit of C tetani is in any way primordial. Both are used to extrude Na+ in exchange for H+ but this is not to drive Na+ energetics, they are much more associated with resistance to high Na+ concentrations and to alkaline pH environments. So it is possible that the N Thermophilus nt-Nha is a relatively modern derivative (it does use a proton tight membrane) of a relatively modern MrpA.
Or, more excitingly, it's possible that an ancestral Na+/H+ antiporter gave rise to both nt-Nha and MrpA. This would be the interesting option as it is possible that the Na+ binding sites, the route across the antiporter for Na+ and the mechanism for activation might just give us the technique used by the original ancestral antiporter. Genetics and structure-function modeling look to be the way to go but I can't see that it's been done yet.
Edit: Found the structure homology studies in here, lying around on my hard drive for years too. End edit.
Peter
Homologous protein subunits from Escherichia coli NADH:quinone oxidoreductase can functionally replace MrpA and MrpD in Bacillus subtilis
In brief they had Bacillus subtilis strains with either an MrpA knockout or an MrpD knockout. The E coli complex I equivalent of NuoL can replace MrpA and the NuoN equivalent can replace MrpD in B subtilis. NouM doesn't seem do either. But all five subunits look very similar to each other and all are clearly related. NuoL, NuoM and NuoN are always described as "antiporter-like". MrpA and MrpD are thought to be antiporters but none ever work alone, the whole complex is needed, so they are probably "antiporter-like" too. They all appear to have been derived from an antiporter but any intrinsic antiporting has been lost.
Which makes me sad because it seems very probable that all of the above subunits are derived from the primordial antiporter at the origin of life which initiated Na+ bioenergetics and all that followed on from that.
Then came Natranaerobius thermophilus. N thermophilus is not really in the league of P furiosus, it's okay growing at up to 57 degC (which will still scald you) but no higher and it has adapted its membrane to remain proton tight at this temperature. BTW it's strictly anaerobic, is an halophile and an alkaliphile. It has (among several) one family of modern antiporters which are clearly genetically related to the MrpA of modern Clostridium tetani (and probably all other MrpAs). Modern nt-Nha is a fully functional antiporter as a stand-alone single gene protein. As these folks say:
The halophilic alkalithermophile Natranaerobius thermophilus adapts to multiple environmental extremes using a large repertoire of Na+(K+)/H+ antiporters
"Gene nt-Nha had 35% identity to the shaA (mrPA) gene of Clostridium tetani. The Mrp proteins belong to the monovalent cation/proton antiporter-3 protein family. This family is composed of multi-component Na+/H+ and K+/H+ antiporters encoded by operons of six or seven genes, and all genes are required for full function in Na+ and alkali resistance (Ito et al., 2000). Sequence analysis of the
regions surrounding gene nt-Nha, however, did not show that it was part of an operon. This indicates that gene nt-Nha does not encode a subunit of an Mrp system, but rather a mono-subunit antiporter".
EDIT number 2: These people have isolated an MrpA from an archaeal species which will antiport on its own, which makes it very similar to nt-Nha. There is also some evidence that complex I can function as a partial Na+/H+ antiporter as in this paper. NuoL is the main suspect. END EDIT.
Neither the MrpA-like nt-Nha nor the modern MrpA subunit of C tetani is in any way primordial. Both are used to extrude Na+ in exchange for H+ but this is not to drive Na+ energetics, they are much more associated with resistance to high Na+ concentrations and to alkaline pH environments. So it is possible that the N Thermophilus nt-Nha is a relatively modern derivative (it does use a proton tight membrane) of a relatively modern MrpA.
Or, more excitingly, it's possible that an ancestral Na+/H+ antiporter gave rise to both nt-Nha and MrpA. This would be the interesting option as it is possible that the Na+ binding sites, the route across the antiporter for Na+ and the mechanism for activation might just give us the technique used by the original ancestral antiporter. Genetics and structure-function modeling look to be the way to go but I can't see that it's been done yet.
Edit: Found the structure homology studies in here, lying around on my hard drive for years too. End edit.
Peter
Monday, March 25, 2019
Life (25) Left or right hand?
Here is the basic Mrp antiporter structure as suggested in
Structure of an Ancient Respiratory System
Quite how many protons are exchanged for how many Na+ is uncertain but there are papers using modern Mrp set in proton tight membranes that suggest it might be more than one H+ in per one Na+ out. ie Mrp is electrogenic, or rather it consumes pH gradient to extrude Na+. This would be no problem in the hydrothermal vent scenario, protons being freely available there. Personally I'd like electroneutrality but that's just my biases as to how P furiosus works. Anyway, Mrp is much like this, with uncertainty about the numbers of ions:
Here is exactly the same antiporter but broken down in to the main channels:
If we ignore the arrows for the H+ in the diagram all we have to do is remove the bulk of MrpA (the N-terminal) and replace it with a power source "pushing" in from the left and we have the membrane bound hydrogenase of P furiosus, still retaining the MrpG Na+ channel and working as a Na+ pump:
The paper then goes on to talk about Complex I and how that, in the Mrp nomenclature, the combined MrpD plus the fused-on C-terminal of MrpA are flipped around. I spent a long time mentally lining up various channels in my head until I twigged the simplest way to look at it was to keep Mrp channels unchanged but look at the NADH dehydrogenase of complex I as simply pumping in to a completely un-flipped Mrp but being bolted on to the opposite end, in the place of the MrpG Na+ channel. Leaving the N-terminal of MrpA still in place, like this:
I've squeezed in an extra MrpD because that's what complex I appears to have done as a modified duplication of either MrpA or MrpD. In mammalian mitochondrial nomenclature the MrpA N-terminal derivative is NuoL, the narrowed (only in this image, not really) MrpD gives NuoM and the full sized MrpD is NuoN. Yu et al only use the bacterial complex I terminology based around the Nqo numbers. I've avoided these numbers (just used the Mrp terminology throughout the doodles) as the switches from terminology to terminology did my head in (as we say here in Norfolk) for weeks. MBH, Mrp and Nqo. Alphabet soup for the subunits!
But the core insight for me was that if you supply power from the left you pump Na+. Supply it from the right and you pump protons. This looks very much like motorising Mrp from one end makes it work in the Na+ extrusion antiport mode. Adding the power source to the opposite end, coincidentally removing the Na+ channel at the same time, drives the antiporter in the H+ expulsion direction, reversing the primordial function of Mrp and so forming the origin of the complex I family.
Obviously there is nothing primordial about complex I. It is reliant on a proton tight membrane and the ability to extract large amounts of energy from NADH, which usually means the presence of molecular oxygen. The least altered representative of antiquity is undoubtedly the MBH of P furiosus and even more so is the ancestor of the Mrp antiporter family.
Peter
Structure of an Ancient Respiratory System
Quite how many protons are exchanged for how many Na+ is uncertain but there are papers using modern Mrp set in proton tight membranes that suggest it might be more than one H+ in per one Na+ out. ie Mrp is electrogenic, or rather it consumes pH gradient to extrude Na+. This would be no problem in the hydrothermal vent scenario, protons being freely available there. Personally I'd like electroneutrality but that's just my biases as to how P furiosus works. Anyway, Mrp is much like this, with uncertainty about the numbers of ions:
Here is exactly the same antiporter but broken down in to the main channels:
If we ignore the arrows for the H+ in the diagram all we have to do is remove the bulk of MrpA (the N-terminal) and replace it with a power source "pushing" in from the left and we have the membrane bound hydrogenase of P furiosus, still retaining the MrpG Na+ channel and working as a Na+ pump:
The paper then goes on to talk about Complex I and how that, in the Mrp nomenclature, the combined MrpD plus the fused-on C-terminal of MrpA are flipped around. I spent a long time mentally lining up various channels in my head until I twigged the simplest way to look at it was to keep Mrp channels unchanged but look at the NADH dehydrogenase of complex I as simply pumping in to a completely un-flipped Mrp but being bolted on to the opposite end, in the place of the MrpG Na+ channel. Leaving the N-terminal of MrpA still in place, like this:
I've squeezed in an extra MrpD because that's what complex I appears to have done as a modified duplication of either MrpA or MrpD. In mammalian mitochondrial nomenclature the MrpA N-terminal derivative is NuoL, the narrowed (only in this image, not really) MrpD gives NuoM and the full sized MrpD is NuoN. Yu et al only use the bacterial complex I terminology based around the Nqo numbers. I've avoided these numbers (just used the Mrp terminology throughout the doodles) as the switches from terminology to terminology did my head in (as we say here in Norfolk) for weeks. MBH, Mrp and Nqo. Alphabet soup for the subunits!
But the core insight for me was that if you supply power from the left you pump Na+. Supply it from the right and you pump protons. This looks very much like motorising Mrp from one end makes it work in the Na+ extrusion antiport mode. Adding the power source to the opposite end, coincidentally removing the Na+ channel at the same time, drives the antiporter in the H+ expulsion direction, reversing the primordial function of Mrp and so forming the origin of the complex I family.
Obviously there is nothing primordial about complex I. It is reliant on a proton tight membrane and the ability to extract large amounts of energy from NADH, which usually means the presence of molecular oxygen. The least altered representative of antiquity is undoubtedly the MBH of P furiosus and even more so is the ancestor of the Mrp antiporter family.
Peter
Sunday, March 24, 2019
Looking to the future
I notice the EU is considering approval for a plan to enforce removal of copyright infringing images form pretty well everywhere on T'internet.
Over many years I've felt that this sort of action would clearly be the end of Hyperlipid as any of us knows it. I back up every month or so to keep my ideas safe for myself but if Hyperlipid suddenly disappears I think everyone should have an idea as to why. This will be it.
Anyway, for now something resembling normal service will be resumed when I can tear myself away from Mrp antiporters and derivatives, which is proving rather difficult.
Peter
Over many years I've felt that this sort of action would clearly be the end of Hyperlipid as any of us knows it. I back up every month or so to keep my ideas safe for myself but if Hyperlipid suddenly disappears I think everyone should have an idea as to why. This will be it.
Anyway, for now something resembling normal service will be resumed when I can tear myself away from Mrp antiporters and derivatives, which is proving rather difficult.
Peter
Friday, March 15, 2019
Tucker on omega six PUFA
While I've been day dreaming about antiporters at the origin of life Tucker has been busy. I guess most people reading here would read Tucker anyway but just in case not, here's the link
Response to Gary Taubes on Omega-6 Fats (Seed Oils) and Obesity
Personally I have no issue with the two concepts being complementary. Carbohydrate in quantities which get glucose past the liver will drive up systemic insulin. Omega six (and 18C omega three) fatty acids will make adipocytes hyper-respond to the obesogenic signal of elevated systemic insulin. Fat (largely dietary sourced) is then lost in to adipocytes. Loss of this fat is the equivalent of not having eaten it, so you are left hungry. It's the old weight gain causes hunger paradigm. I like it.
Peter
Response to Gary Taubes on Omega-6 Fats (Seed Oils) and Obesity
Personally I have no issue with the two concepts being complementary. Carbohydrate in quantities which get glucose past the liver will drive up systemic insulin. Omega six (and 18C omega three) fatty acids will make adipocytes hyper-respond to the obesogenic signal of elevated systemic insulin. Fat (largely dietary sourced) is then lost in to adipocytes. Loss of this fat is the equivalent of not having eaten it, so you are left hungry. It's the old weight gain causes hunger paradigm. I like it.
Peter
Wednesday, March 06, 2019
Life (24) Porting over CCCP
Just to summarise: The membrane bound hydrogenase (MBH) of Pyrococcus furiosus pushes a proton outwards through a proton permeable membrane which then returns through a subunit of MBH which looks very much like three quarters of the Mrp antiporter. The Mrp antiporter is derived from a very, very ancient antiporter which appears to have been one of the core systems of the last universal common ancestor (LUCA). Which ion travels in which direction in the modern families of Mrp is currently not particularly clear and may depend on all sorts of factors. OK.
No one has worked out the detailed structure/function of the Mrp antiporter, but there is a very interesting paper from back in 2001 which might give us some insight about function at least.
Mrp‐dependent Na+/H+ antiporters of Bacillus exhibit characteristics that are unanticipated for completely secondary active transporters
The study used modern E coli whose plasma membrane is tight to both protons and Na+. A strain with all of its antiporters deleted was used and then plasmids were engineered to supply a single gene antiporter of the nahA type (also from E coli) or an Mrp from Bacillus pseudofirmus OF4 (yup, that's its name) or one from Bacillus subtilis.
Respectively we have strains ending in -118 endowed with the blank plasmid, -nhaA for the monogenic antiporter, -BSmrp (B subtilis Mrp) or -OFmrp (B pseudofirmus Mrp). They stuck the engineered E coli in to 25mmol of NaCl, fed it and looked at the intracellular Na+ concentration. Very simple.
The first two columns are exactly what you would expect. Having no antiporter gives an intracellular Na+ in equilibrium with culture medium Na+ at around 25mmol, near enough. Any antiporter rescues this, the nhaA monogenic antiporter somewhat better than either of the Mrp antiporters.
Now here comes the clever bit. They added CCCP, a classical protonophore which drops the membrane potential from 150-ish mV to 15-ish mV (obviously the membrane voltage should be zero but the E coli complex I will be working flat out to stop this fatal occurrence plus I suspect the dose of CCCP used was less than supramaximal, though I've not checked this), and then they looked at the intracellular Na+ concentration. So they have suddenly converted a modern E coli to an E coli with a proton permeable, Na+ impermeable cell membrane. Like Pyrococcus but without the boiling water. Or like LUCA. Both organisms to which Mrp antiporting is/was very important.
The simple nhaA is utterly dependent on a proton tight membrane with a transmembrane proton gradient and it fails to antiport anything in the presence of CCCP, as you would expect (line two, both right hand columns). The nhaA is not primordial.
Both of the Mrp type antiporters maintain an intracellular Na+ between 13.4mmol and 16.9mmol, which is "high-but-physiological", using a CCCP proton "leaky" membrane voltage of 15 mV to effectively pump out Na+.
Quite how the Mrp antiporters do this is unclear. Most of the work has been done on the big subunits, A and D. One is probably a proton channel and one a Na+ channel but even this is not completely clear and in Yu et al's Structure of an Ancient Respiratory System they consider both MrpA and D to have proton channels. So it's messy. The combined small subunits, MrpE, MrpF and MrpG, plus the tail end of MrpA appear to be a proton/Na+ antiporter in their own right. I'll refer to this section as a "simple" antiporter.
Clearly, trying to get a Na+ gradient from a proton leaky membrane by making use of a monogenic nhaA type antiporter doesn't work. Using an Mrp antiporter does.
OK, wild speculation time.
I consider the arrangement in Pyrococcus furiosus is necessary because the cell membrane is permeable to protons. Pump a proton outwards and generally it will boomerang back. Pump it directly in to the mouth of an antiporter and it will return while antiporting a Na+ outwards. The Na+ stays outside. It does this using much of the Mrp machinery.
The end game is to drop a precious proton down the throat of the simple antiporter, without losing it through a leaky membrane. I think the Pyrococcus MBH keeps this proton "in-complex" to avoid losing it. Power is probably supplied to the left hand proton channel from the FeNi hydrogenase by the loop cluster and helix HL.
Like this, yellow circles are antiporters:
Looking at Mrp, I think it is the precursor of the MBH and is doing exactly the same thing but geochemically, ie it is an adaptation to a low geochemical proton gradient across a proton leaky membrane. It still takes the proton from a 15mV proton motive force but it initially uses this to antiport another proton outwards, protects this one from loss through the leaky membrane by keeping it "in-complex" and uses this to antiport Na+ outwards, which stays outside:
In both diagrams everything is identical to the right of the FeNi hydrogenase or MrpA N-terminal domain. All that differs is the method for "elevating" the guarded proton to the entrance of the simple antiporter on the right.
TLDR: Mrp is an adaptation to a failing geochemical proton gradient. Membrane bound hydrogenase is the adaptation to a failed geochemical gradient.
It is amazing to me that Mrp keeps its core function today despite the radically different tasks (saline and alkaline tolerance) which it is needed for nowadays. All we need is a proton leaky membrane to return it to do what it did initially. No change.
Okay, it's not an envelope, its the back of a chocolate wrapper. Speculation is such fun, given enough chocolate.
Peter
No one has worked out the detailed structure/function of the Mrp antiporter, but there is a very interesting paper from back in 2001 which might give us some insight about function at least.
Mrp‐dependent Na+/H+ antiporters of Bacillus exhibit characteristics that are unanticipated for completely secondary active transporters
The study used modern E coli whose plasma membrane is tight to both protons and Na+. A strain with all of its antiporters deleted was used and then plasmids were engineered to supply a single gene antiporter of the nahA type (also from E coli) or an Mrp from Bacillus pseudofirmus OF4 (yup, that's its name) or one from Bacillus subtilis.
Respectively we have strains ending in -118 endowed with the blank plasmid, -nhaA for the monogenic antiporter, -BSmrp (B subtilis Mrp) or -OFmrp (B pseudofirmus Mrp). They stuck the engineered E coli in to 25mmol of NaCl, fed it and looked at the intracellular Na+ concentration. Very simple.
The first two columns are exactly what you would expect. Having no antiporter gives an intracellular Na+ in equilibrium with culture medium Na+ at around 25mmol, near enough. Any antiporter rescues this, the nhaA monogenic antiporter somewhat better than either of the Mrp antiporters.
Now here comes the clever bit. They added CCCP, a classical protonophore which drops the membrane potential from 150-ish mV to 15-ish mV (obviously the membrane voltage should be zero but the E coli complex I will be working flat out to stop this fatal occurrence plus I suspect the dose of CCCP used was less than supramaximal, though I've not checked this), and then they looked at the intracellular Na+ concentration. So they have suddenly converted a modern E coli to an E coli with a proton permeable, Na+ impermeable cell membrane. Like Pyrococcus but without the boiling water. Or like LUCA. Both organisms to which Mrp antiporting is/was very important.
The simple nhaA is utterly dependent on a proton tight membrane with a transmembrane proton gradient and it fails to antiport anything in the presence of CCCP, as you would expect (line two, both right hand columns). The nhaA is not primordial.
Both of the Mrp type antiporters maintain an intracellular Na+ between 13.4mmol and 16.9mmol, which is "high-but-physiological", using a CCCP proton "leaky" membrane voltage of 15 mV to effectively pump out Na+.
Quite how the Mrp antiporters do this is unclear. Most of the work has been done on the big subunits, A and D. One is probably a proton channel and one a Na+ channel but even this is not completely clear and in Yu et al's Structure of an Ancient Respiratory System they consider both MrpA and D to have proton channels. So it's messy. The combined small subunits, MrpE, MrpF and MrpG, plus the tail end of MrpA appear to be a proton/Na+ antiporter in their own right. I'll refer to this section as a "simple" antiporter.
Clearly, trying to get a Na+ gradient from a proton leaky membrane by making use of a monogenic nhaA type antiporter doesn't work. Using an Mrp antiporter does.
OK, wild speculation time.
I consider the arrangement in Pyrococcus furiosus is necessary because the cell membrane is permeable to protons. Pump a proton outwards and generally it will boomerang back. Pump it directly in to the mouth of an antiporter and it will return while antiporting a Na+ outwards. The Na+ stays outside. It does this using much of the Mrp machinery.
The end game is to drop a precious proton down the throat of the simple antiporter, without losing it through a leaky membrane. I think the Pyrococcus MBH keeps this proton "in-complex" to avoid losing it. Power is probably supplied to the left hand proton channel from the FeNi hydrogenase by the loop cluster and helix HL.
Like this, yellow circles are antiporters:
Looking at Mrp, I think it is the precursor of the MBH and is doing exactly the same thing but geochemically, ie it is an adaptation to a low geochemical proton gradient across a proton leaky membrane. It still takes the proton from a 15mV proton motive force but it initially uses this to antiport another proton outwards, protects this one from loss through the leaky membrane by keeping it "in-complex" and uses this to antiport Na+ outwards, which stays outside:
In both diagrams everything is identical to the right of the FeNi hydrogenase or MrpA N-terminal domain. All that differs is the method for "elevating" the guarded proton to the entrance of the simple antiporter on the right.
TLDR: Mrp is an adaptation to a failing geochemical proton gradient. Membrane bound hydrogenase is the adaptation to a failed geochemical gradient.
It is amazing to me that Mrp keeps its core function today despite the radically different tasks (saline and alkaline tolerance) which it is needed for nowadays. All we need is a proton leaky membrane to return it to do what it did initially. No change.
Okay, it's not an envelope, its the back of a chocolate wrapper. Speculation is such fun, given enough chocolate.
Peter
Life (23) Antiporting: Choose your ion well
Sorry to go on about Yu et al's paper
Structure of an Ancient Respiratory System
but there's an awful lot in it. Here again is their working model of the MBH from Pyrococcus furiosus:
The Na+ channel on the right hand end shown as MbhC was looked at in great detail in the paper and is very convincingly a Na+ channel. The group never looked at the actual structure of the unpowered antiporter Mrp so they are working with other people's assumptions, some of which are quite likely better than others as regards the nature of the ion favoured by a particular channel. So here is the membrane arm of MBH they ended up with, labelled to represent the unpowered original Mrp antiporter. Obviously the hydrogenase has been replaced by the MrpA N-terminal:
Now the problem with this, apart from the highly electrogenic charge exchange of three protons in for one Na+ out, is that the left hand MrpA module is actually a Na+ channel. There's lots of pretty convincing evidence for this but I won't waffle on any more about it. So lets set up the Mrp antiporter as a pure H+/Na+ exchanger like this:
Two protons inwards, two Na+ antiported outwards, what could be nicer? This is the structure favoured by group in Sweden comparing MrpA and MrpD with the antiporter-like subunits of complex I.
Functional Differentiation of Antiporter-Like Polypeptides in Complex I; a Site-Directed Mutagenesis Study of Residues Conserved in MrpA and NuoL but Not in MrpD, NuoM, and NuoN
I think it might be completely wrong.
I like this paper but I'm still left with a niggling thought that having two antiporters in one complex, apparently both doing the same thing, is just a little wasteful. The normal bacterial approach is one where the jettisoning of genes is developed to a fine art.
So Yu et al and Sperling et al have rather different ideas about the function of various channels in the Mrp complex. No one is really talking about why there should be four channels...
The modern Mrp antiporter is very interesting in it's own right. It needs a little post of its own.
Peter
Structure of an Ancient Respiratory System
but there's an awful lot in it. Here again is their working model of the MBH from Pyrococcus furiosus:
The Na+ channel on the right hand end shown as MbhC was looked at in great detail in the paper and is very convincingly a Na+ channel. The group never looked at the actual structure of the unpowered antiporter Mrp so they are working with other people's assumptions, some of which are quite likely better than others as regards the nature of the ion favoured by a particular channel. So here is the membrane arm of MBH they ended up with, labelled to represent the unpowered original Mrp antiporter. Obviously the hydrogenase has been replaced by the MrpA N-terminal:
Now the problem with this, apart from the highly electrogenic charge exchange of three protons in for one Na+ out, is that the left hand MrpA module is actually a Na+ channel. There's lots of pretty convincing evidence for this but I won't waffle on any more about it. So lets set up the Mrp antiporter as a pure H+/Na+ exchanger like this:
Two protons inwards, two Na+ antiported outwards, what could be nicer? This is the structure favoured by group in Sweden comparing MrpA and MrpD with the antiporter-like subunits of complex I.
Functional Differentiation of Antiporter-Like Polypeptides in Complex I; a Site-Directed Mutagenesis Study of Residues Conserved in MrpA and NuoL but Not in MrpD, NuoM, and NuoN
I think it might be completely wrong.
So Yu et al and Sperling et al have rather different ideas about the function of various channels in the Mrp complex. No one is really talking about why there should be four channels...
The modern Mrp antiporter is very interesting in it's own right. It needs a little post of its own.
Peter