Okay. Time to finish the complex I series. Under conditions of a cell surface membrane which is partially permeable to protons and (less so) to hydroxyl ions there can be a proto-metabolism based on the ingress of protons driving both carbon fixation and energy generation, with neutralisation by OH
- ions. This is dependent on having a partially permeable membrane to both of these ions. Subsequently, by using the simultaneous impermeability to (larger, less permeant) Na
+ ions, combined with the above ability to neutralise protons with OH
-, a Na
+/H
+ antiporter can establish a Na
+ potential to drive a proto-ATP synthase. Koonin's group discussed it here:
Evolutionary primacy of sodium bioenergeticsAs the protocell membrane becomes progressively more impermeable to both H
+ and OH
- then running a Na
+/H
+ antiporter becomes progressively more difficult. At the same time this makes proton pumping potentially advantageous. This is how I am guessing that proton pumping may have developed.
If we start from that neat doodle from
looking like this:
we can reverse model it back to a simpler NiFe hydrogenase in a proton semi-permeable membrane and need just four images to sum it up:
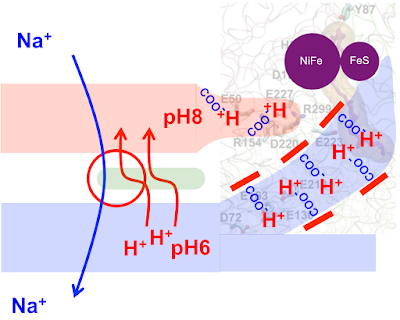
This has the ocean at pH 6 protonating acidic amino acids in a channel from the ocean to the FeS cluster. There is also a side chain of acidic amino acids in contact with the NiFe cluster which are non-protonated because they are contiguous with the cytoplasmic fluid of pH10.
A molecule of hydrogen arrives at the NiFe cluster and is split in to a pair of electrons and a pair of protons:
The electrons hop on to the FeS cluster and thence to ferredoxin (accompanied by their ability to do work) to give reduced ferredoxin, Fd2-, while the protons go to the waiting carboxylates of the amino acids on the route to the pH10 cytoplasm:
which then leaves the complex ready for the next hydrogen molecule to come along after the protons on the cytoplasmic route's amino acids have been deprotonated by the pH10 cytoplasm:
The cost of this manoeuvre being a small fall in the intracellular pH, to be neutralised by the same alkaline vent fluid which supplied the molecular hydrogen.
That seems quite simple.
If we consider what might happen if the availability of hydroxyl ions is curtailed by progressively rising impermeability of the cell membrane to both H+ and OH- then the process must halt. With the evolution of soluble hydrogenases, and especially of electron bifurcation, then Fd2- might become more plentiful but molecular hydrogen less so.
We can consider what the immediate advantage might be to a cell to consume Fd2- and regenerate molecular hydrogen by running this complex in reverse.
So now I've set the intracellular pH to pH7 and left the ocean fixed at pH6. It's a big ocean.
In this scenario all of the amino acids in the complex would be protonated:
If we allow a Fd2- molecule to place a pair of electrons on to the FeS cluster:
these can combine with a pair of protons to form molecular hydrogen. These protons should come from the (very slightly) more acidic ocean channel:
This reaction is exothermic and needs no proton gradient. It leaves us with a deficit of protons in the oceanic pH channel:
which you would expect to be replenished from the bulk ocean. But we have a certain amount of free energy available from the high energy Fd2- molecule used to make the molecular hydrogen. All that is needed is an electrostatic/conformational change comparable to the "Doohickey" function of the last several posts and it becomes simple to take two protons from the cytoplasmic influenced amino acids and put then on to the oceanic side using the energy available from Fd2- oxidation:
What might be the immediate advantage of doing this?
The pH7 environment on the cellular side will allow spontaneous re-protonation of the acidic residues in the complex:
which will clearly leave a very small and very localised area of higher pH, here designated as pH8 for illustrative purposes only:

We have now produced a very localised accentuation of the progressively feebler pH gradient resulting from the cell membrane becoming progressively more opaque to OH- ions.
As cell energetics are highly Na+ dependent, as per the introduction to this post, establishing a small area of accentuated pH gradient will allow the immediate advantage of facilitating the struggling Na+/H+ antiporting process, at the cost of allowing the loss of the newly developed localised area of pH 8 (as shown) back down to pH7 (not shown). Like this:
which is fine except a simple "monogenetic" antiporter is actually pretty useless at low membrane potentials, as in:
So it would be better to have the ancient ancestor of the modern MRP ultra-low proton gradient antiporter instead. Here we have several protons each "kicking" another inward channel to finally
"kick" a Na+ ion out of the cell:
At this point having MRP snuggle up to a membrane bound hydrogenase to access a better pH gradient is starting to look vaguely like a complex I precursor, but not quite. All we have is a small improved localised pH gradient, no gross expulsion of protons, and the sole use is to generate a Na+ ion gradient. But that would be advantageous, immediately.
Now let's worsen matters still further and drop the intracellular pH to 6.5, where even the mighty MRP antiporter is in trouble. We can get extracellular protons to the half way inward mark, and intracellular Na+ to the half way outward mark but there is insufficient pH drive to complete their respective journeys. Stalemate:
Now if we just think about that energy input from "wasting" a Fd2- molecule we can have a conformational/electrostatic change in the green outlined amino acid (modern day aspartate D72 in the original diagram) like this:
giving a "push" to help the struggling MRP antiporter:
by providing a "kick" which the pH gradient can't manage alone. As it stands there need be no outward proton translocation, just a push to the MRP antiporter. In fact the localised pH gradient would be lost on Na+ antiporting but the cell would have bought a better Na+ gradient for ATP synthase in return:
In this last image I've suggested that blocking off access to the ocean would be an incremental advantage too as it might make a conformational change in the "kicking" acidic amino acid more effective at facilitating MRP antiport completion.
None of this is a proton pump. But as the cell membrane become essentially impermeable to protons there develops an advantage to running MRP in reverse. All you have to do is attach the kicking-complex the wrong way round to MRP and you could kick a Na+ in to the cell and two H+ out of the cell, then start of using protons in ATP synthase. Or completely drop the module which translocates Na+ and just use the "kick" to push two protons outwards. Or, given a power source like the NADH:CoQ couple, kick four protons out wards, as in complex I. Notice the "kicker" is on the opposite end of the MRP antiporter derivative here and the Na+ module has been abandoned/replaced:
Given a less potent power source such as the Fd2-/H+ couple you can just drive out one proton, as Ech does:
You can also, if you're Pyrococcus furiousus living at 100degC, still pump Na+ ions (it's not easy to build a proton tight membrane at 100degC, so Na+ energetics are retained) by flipping one proton channel round, pushing a proton outwards and allowing this proton back inwards to antiport a Na+ ion outwards:
which is a proton-neutral technique to establish a Na+ ion gradient.
All you have to do is to develop a "kicker" for MRP and the world is your oyster. There are many derivatives of this type of pump with various subunits arranged in various orders. It's a molecular Lego set. All that is needed is for each step during its development to be continuously advantageous.
The concept that modern derivatives might be the best guide as to where and how life began fascinates me and has been laid out by Nick Lane's group here:
It makes a lot of sense to me.
Peter