The people who wrote this review are plant biologists. Nothing wrong with that and they've been doing some fairly deep thinking about reactive oxygen species signalling.
In this paper I'd like to just run through the images in Fig 1 from Box 1 with some sort of commentary.
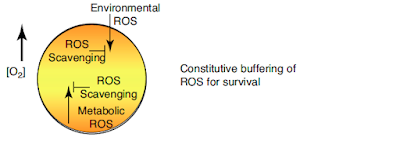
This is an ancestral organism. It could represent LUCA, which has implications about what LUCA might have needed to do to deal with ROS. Under conditions of rising oxygen tension (or even without rising oxygen tension) there is the need for basic ROS scavenging simply to survive. Any organism which couldn't do this has not left descendants:
If you want to maximise your chances of survival then the co-opting of some sort of protein as an ROS receptor provides an advantage. A better controlled response to suppress ROS to acceptable levels becomes possible without wasting resources on excessive scavenging systems. The little horse-shoes are the receptors:
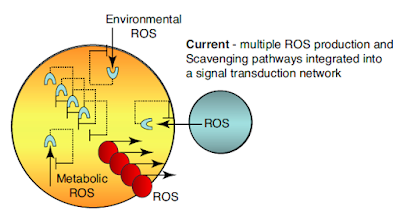
This review is from 2015, at which time ROS generation by bacteria was thought to be limited to "accidental" release during photosynthesis by cyanobacteria. Obviously this is no longer the case but anyway, we can think of the blue circle as representing an organism or cell as part of the same organism capable of generating and releasing significant ROS, which our golden organism might not like and might dedicate another sensing/quenching system to:
If the sensing system turns out to be carrying useful information about the neighbouring organism/cell our golden cell might consider it worth the NADPH to send a signal back. Maybe a "come on" signal, maybe a "bugger off" signal. But ROS based too. Here we are talking eukaryotes as the ROS generator (in this review) is considered to be one of the NADPH oxidases (NOX), the small red bauble, which does not appear to be coded for in prokaryote genomes:
Assuming that cell-cell signalling proves helpful, the system can go on to develop as many responses as is useful to survival:
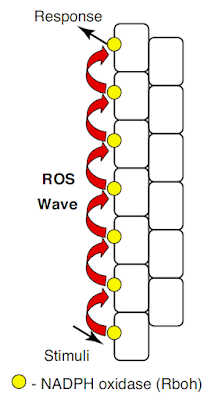
Back to plant biology. This next image summarises what the group thinks happens in groups of plant cells, this time taken from Figure 1 of the text proper:
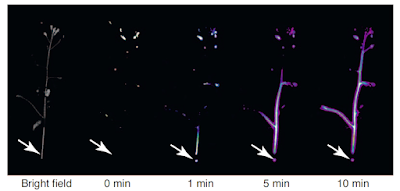
And because they are plant biologists, and because plants sit still (on a human timescale) when you kick them, here is what happens when you "hurt" or injure an Arabidopsis plant, illustrated using an fluorescence probe. There is a luciferase gene spliced in to the ROS generating machinery. Note the timescales:
So plants appear to signal within the organism, on a timescale which is appropriate to their needs, using ROS and ROS receptors. I've been curious about how an oak tree root hair might signal to the top of the canopy. Even for a plant speed of communication needing several days might be too slow.
Is it possible to convert a localised ROS signal to an organism-wide information network signal?
Maybe.
Peter