Before trying to work backwards from complex I via NiFe hydrogenases to consider how such a system might have evolved I've spent a couple of weeks on Fig 6 of this paper:
The coupling mechanism of respiratory complex I — A structural and evolutionary perspective
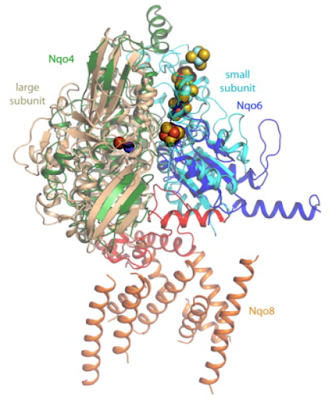
For orientation this is section A from Fig 1 in the same paper:
The coupling mechanism of respiratory complex I — A structural and evolutionary perspective
For orientation this is section A from Fig 1 in the same paper:
and most of the rest of the post is going to be about the area outlined by the red oval.
Fig 4 is an overlay of this area taken from complex I with the same area from the NiFe hydrogenase of the sulphate reducing bacterium Desulfovibrio gigas. Both of these complexes show marked conservation of their structures in this area, as do all similar complexes:
and if I duplicate the image I can add in the electron pathways like this:
In this next image I've put in both pathways and a funny little white, green and blue doo-hickey which converts the energy of the reaction in to a conformational change in the protein responsible for driving ion translocation. The doo-hickey is what Fig 6 is going to look at in detail. I chose the colour scheme here to match that in Fig 6:
This is the un-altered original Fig 6. I'll explain it step by step in the following section:
In my modified images a solid red circle is a full electron negative charge. Open circles in black and white represent charge distributions of the magnitude used for hydrogen bonding, either +ve or -ve as appropriate. A solid red cicle with a +ve is a proton.
Part 1 can be thought of as the open configuration. Electrons have arrived down the FeS chain from NADH to the terminal clusters N2 and (probably) N6a. Their negative charge induces a change in the yellow protein which, in combination with hydrogen "repulsion" between key amino acids opens the binding site to allow CoQ to enter. The original image had just two small + signs, within black circles, to designate mildly positively charged amino acid residues on the "top" end of the green protein. The same mild positive charge is also present on amino acids X and Y of the blue protein but were missing from the original image so I've added them in. These mildly positive areas are doing the opposite of hydrogen bonding, what I called hydrogen repulsion. The proteins are held apart, that's what the purplish thick arrows signify:
Now oxidised CoQ docks with its binding site. Note that the pair of keto oxygens on CoQ are mildly -ve charged so form normal attractive hydrogen bonds to the mildly +ve amino acid residues X and Y on the blue protein. I've put in the partial charges eliciting the normal hydrogen bonding which are absent from the original image using hydrogen bonding -ves within small open circles:
The two electrons transfer from their FeS clusters to CoQ to give a strongly negatively charged oxygen atom on either side of CoQ, call it CoQ2-. I've used the same red circles to accentuate the small negative signs used in the original diagram:
These strongly negatively charged oxygen atoms exert a pull (dark purple arrows pointing together) which induces the conformational change in to the location of the green protein which causes the actual pumping. This is shown in section 4 where the green protein has moved "upwards" and pumping has happened. In the same process the exit of the electrons from the FeS clusters allows the yellow protein back in to its starting conformation:
The next change is subtle. All that has happened is that CoQ2- has taken the protons from the amino acids X and Y to form neutral CoQ2H, leaving the amino acids with the full negative charges. The covalent bonds rearrange slightly and the red -ve circle charges move a tiny distance on to the blue protein:
At this point CoQ2H is electrically neutral but still held in place by hyrdogen bonding to the now negatively charged amino acids X and Y, as are the two mildly positively charged amino acids (black +ves in white circles) on the green protein.
Two things have to happen to return the complex to the section 1 configuration. Protons must enter the active site to allow the amino acids X and Y to lose their net negative charges and electrons must be replaced in the FeS clusters N2 and N6a to allow the yellow protein to assume the "open" configuration as CoQH2 exits:
Once these two events have occurred then we are back in to the situation in section 1 and the process can repeat:
The system is reversible and, given an adequate membrane potential, conformational changes can result in electron translocation in reverse up to the flavin unit where they can be donated to NAD+ to give NADH. Or to oxygen to give superoxide.
I think it might be worth taking a pause here before looking at the possible mechanism involved in the NiFe hydrogenase mechanism of action. As the authors comment:
"A similar scheme can be used for the mechanism of NiFe-hydrogenases, where charge variations in the NiFe site drive conformational changes resulting in proton translocation."
Clearly there is nothing ancestral about the CoQ system but it's mechanism of action has to be derived from that of the NiFe hydrogenases, which do appear to carry the signature of the ancestral pumping mechanism.
Peter











3 comments:
OK, this is a REALLY dumb question. It's something I've been wondering about for a while.
If one gets up close and personal with a molecule, does it really look like curly tinsel?
Haha, probably not. More deeply you are questioning how we might "visualise" something for which photons are large particles of energy rather than anything which might form an image...
It's like trying to define the temperature of a water molecule embedded in an complex protein. Sure, it vibrates more if it's "hotter", but how does this translate to infra red radiating emission we feel as temperature?
But the ribbon doodles are very pretty!
Peter
Well, yes, when I said, "look like" I was speaking metaphorically.
Yes, the ribbon doodles are pretty, but I find the old style tinkertoy representation a lot easier to understand! No doubt the ribbons are more accurately representational, for whatever that might mean at that scale, because biology is messy 😜
Post a Comment